Analysis
In comparative experiments, statistical analysis is used to estimate effect size and determine the weight of evidence against the null hypothesis.
Content:
- Method of analysis
- Parametric and non-parametric tests
- Data transformation
- Multiple testing corrections
- Post-hoc testing
- Number of factors of interest
In the EDA diagram, an analysis node should receive input from at least one outcome measure and at least one variable of interest should be included as a factor of interest in the analysis.
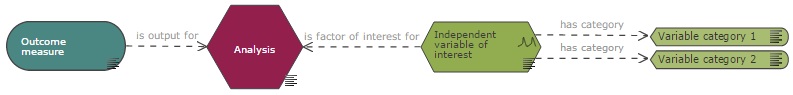
If nuisance variables are included in the analysis, for example as blocking factors or covariates, then they should be connected to the analysis node, using the appropriate link.
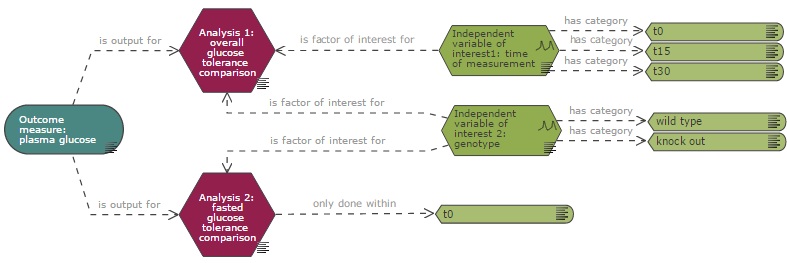
In a situation where only a subset of the recorded data is included in the analysis, for example if an analysis is carried out at a selected time point (see analysis 2 in the example below) when responses were measured at several time points, the variable category (and not the variable itself) should be connected directly to the analysis node to indicate that the analysis is only conducted on the data from that category.
Information to be provided in the properties of the analysis node include the method of analysis, whether a post-hoc test and multiple testing corrections are used and whether the analysis was conducted blind (see blinding section). In case blinding was not achieved, the reason why should also be specified.
If multiple analyses are performed on the experimental data, one analysis node per analysis should be included on the diagram. The primary analysis, which is used to calculate the sample size should be identified.
Method of analysis
Statistical analysis strategies should be chosen carefully to ensure that valid conclusions are drawn from the data. Choice of an appropriate test depends on the number of outcome measures, the properties of these outcome measures, the independent variables of interest and whether any additional nuisance sources of variability (i.e. nuisance variables) need to be accounted for in the analysis. It also depends on whether or not the data collected satisfies the parametric assumptions.
Once the diagram of an experiment is complete and the feedback from the critique has been dealt with, the system can generate a suggestion of statistical tests which are compatible with the design of the experiment. At this stage, the system has no information on whether the data satisfies the parametric assumptions required when performing a parametric analysis and the user has to assess these assumptions, for example checking the data is normally distributed, in order to choose which method to use.
Before the experiment is carried out, a planned method of analysis can be selected from the drop down menu in the properties of the analysis node. Once the data are collected, there is sometimes a rationale to deviate from the planned method. For example, the data does not satisfy the parametric assumptions and data transformation fails to address this, thus a non-parametric analysis method appears to be more appropriate. In such a situation, the diagram can be updated and the method of analysis used, along with the reason for not using the planned method can be indicated.
Regardless of whether the data satisfy parametric assumptions, how these assumptions are assessed should be indicated in the properties of the analysis node.
Parametric and non-parametric tests
Parametric tests have more statistical power than non-parametric tests, as long as the parametric assumptions are met. When the data does not satisfy these assumptions, non-parametric tests are more powerful. However it is important to note that in many cases the parametric assumptions can be satisfied by utilising a data transformation. This should be attempted before a non-parametric method of data analysis is considered.
When assessing the assumptions, it should be noted that the residuals of the analysis, as well as the data itself, needs to be considered. For each observation in the dataset:
observation = predicted + residual
where the ‘predicted value’ is the value predicted by fitting the statistical model to the data (for example a group mean) and the ‘residual’ is the difference between the observation and its prediction.
Data are considered to be suited to parametric analysis if they satisfy the following assumptions:
- The data should be continuous, rather than categorical or binary
- The responses should be independent i.e. each observation should not be influenced by any other, once all sources of variability are accounted for.
- The residuals from the analysis should be normally distributed
- The different groups have similar variances (the homogeneity of variance assumption)
Normal distribution
Parametric tests, such as the t-test, ANOVA and ANCOVA, carry the assumption that your responses, or more precisely the residuals from the analysis, are approximately normally distributed. Many biological responses follow this distribution. If a response that is normally distributed is measured repeatedly, under the same experimental conditions, you would expect most of the responses to lie close to the centre of the distribution, with increasingly fewer responses observed as you move away from the centre. There will be approximately the same number of responses observed above the centre as below it, giving a symmetric distribution.
There are various tests for normality, for example the Shapiro-Wilk and Kolmogorov-Smirnov tests. However these tests struggle to detect non-normality if the sample size is small, as is often the case with animal experiments. An alternative approach is to produce a normal probability plot. This involves plotting the residuals (each observation minus the mean of the group) against what they would be if they were normally distributed. If the points on the plot lie along a straight line, then this is a good indication that the normality assumption holds.
For more information on producing a normal probability plot, click here.
Homogeneity of variance
This is one of the key assumptions of most parametric tests and it implies that the variability of the response is not dependent on the size of the response. For example, the within-group variability does not depend on the size of the group mean. This is because the null hypothesis being tested is that the groups are samples from the same population and so any differences in the means, or variances, are due to chance.
With biological responses, the variability often increases in magnitude as the response increases. In this case the assumption of homogeneity of variance may not hold.
There are several tests that can be used to assess the homogeneity of variance, including the Brown-Forsythe test and Levene’s test. However, as with the formal tests of normality these are not recommended when the sample size is small, as is often the case with animal experiments. An alternative approach is to produce a residual vs. predicted plot. If the scatter of points on this plot is random, with no patterns such as a fanning effect (indicating the residuals get larger as the predicted values increase) then the homogeneity of variance assumption holds. More information on producing a residual vs. predicted plot can be found in Bate and Clark (2014).
Data transformation
If the data are not normally-distributed and/or the homogeneity of variance assumption does not hold, a data transformation can help the investigator satisfy the parametric assumptions and hence enable them to use the more sensitive parametric tests. Data transformation involves replacing the variable with a function of that variable.
If the outcome measure is transformed, this can be indicated by connecting it to a data transformation node. Data transformation nodes are outcome-specific; two different outcome measures cannot be connected to the same data transformation node even if both outcomes are transformed the same way. The data transformation produces the output which is used in the analysis.

Common transformations include ‘mathematical’ transformations (e.g. log, square root or arcsine) or the rank transformation:
Loge and log10 transformation
This transformation is useful when a response increases exponentially (for example bacterial cell counts). It is perhaps the most common transformation applied to biological data and involves taking the log value of each observation. This can be performed on either the log10 or loge scale.
One disadvantage of applying a log transformation is that zero or negative responses cannot be transformed. If the outcome measure being analysed contains zero or negative values, it is necessary to add a small offset onto all responses such that all data are positive.
Square root transformation
Square root transformation involves taking the square root of each response. A disadvantage of the square root transformation is it cannot be applied to negative numbers. Therefore if any responses are negative, a constant should be added to all numbers to ensure they are all positive.
If the response is a count response then it is worth considering a square root transformation prior to analysis.
Arcsine transformation
This consists of taking the arcsine of the square root of a number. For proportion responses, which are bounded above by 1 and below by 0, an arcsine transformation may be appropriate. With responses that are bounded above and below, there is a tendency for the variability of the response to decrease as it approaches these boundaries. The arcsine transformation effectively increases the variability of the responses at the boundaries, and decreases the variability of the responses in the middle of the range.
Any response that is bounded above and below can be transformed using this method, but if the responses are not contained within the range of 0 to 1, then it will need to be scaled to fit this range first.
Rank transformation
When the parametric assumptions do not hold, even after a mathematical transformation, a rank transformation can be used so that the parametric tests can be applied (to rank transformed data).
A rank transformation consists in ranking the responses in order of size, with the largest observation given the rank 1, the second largest, rank 2, and so on.
Note that rank transforming the data loses information and potentially reduces the power of the experiment. The ranking technique also implies that the results of the statistical analysis are less likely to be influenced by any outliers. For example, on the original scale, the largest observation in the dataset may appear to be an outlier, but it is given rank 1 regardless of the actual numerical value. This observation will be given the same rank regardless of how extreme it is.
More information on these transformations can be found in Bland and Altman (1996), Conover and Iman (1981) and Bate and Clark (2014). All above-mentioned transformations can be applied when using InVivoStat and are available from the ‘Transformation’ drop-down menu within the parametric analysis modules.
Multiple testing corrections
In many biology-related fields it is relatively common to test multiple hypotheses and hence make multiple comparisons in a single experiment. For example, if several behaviours are measured, or the experiment is high throughput or a genomics/proteomics experiment. In such situations, it is important to correct for random events that falsely appear to be significant.
In an experiment with one hypothesis, a significance level (α) set at 0.05 means that when the null hypothesis is true the probability of obtaining a true negative result is 95% and the probability of obtaining a significant result by chance (a false positive) is 5% (1 – 0.95).
With the same significance level, for an experiment with 3 hypotheses the probability of obtaining a false positive result is raised to 14% (1 – 0.953), with 25 hypotheses it is raised to 72% (1 – 0.9525), and with 50 hypotheses, to 92% (1 – 0.9550). The aim of multiple comparison procedures is to reduce this probability back down to 5%. See the Bonferroni correction described below.
Post-hoc testing
If an experiment is analysed using an ANOVA approach and the null hypothesis is rejected, this implies that there is a difference among the group means but it does not indicate which groups are different. To determine where the differences lie specifically, subsequent post-hoc testing can be performed.
There are several post-hoc tests to choose from, depending on whether the post-hoc comparisons were planned in advance or not and whether all-pairwise comparisons, or only comparisons with the control group are required. Only one post-hoc test should be run per experiment.
Tests adjusted for multiple comparisons
Dunnett test
The Dunnett test is a good choice when comparing groups against a control group and it provides a correction for multiple comparisons, thus reducing the likelihood of false positive conclusions (type I error).
Tukey HSD (honest significant difference)
The Tukey HSD test is useful when comparing everything with everything. It calculates all possible pairwise comparisons and provides adequate compensation for multiple comparisons. However for selected pairwise comparisons, this post-hoc test is overly strict and therefore increases the likelihood of false negative conclusions (type II error).
Bonferroni test
The Bonferroni test can be used for planned comparisons between a subset of groups, and concurrently adjusts the significance levels to account for multiple comparisons. This test is considered highly conservative and is thus associated with a higher false negative (type II error) rate.
To perform the Bonferroni correction, the significance level is readjusted by dividing it by the number of comparisons. Thus for 3 comparisons, the significance cut-off point is set at 0.017 (α/n = 0.05/3), and for 25 comparisons, it is set at 0.002 (α/n = 0.05/25). This correction is simple to perform but very conservative; it greatly reduces the risk of obtaining false positives but increases the risk of obtaining false negative when dealing with large numbers of comparisons.
Step-wise tests
Alongside the tests above, all of which apply a single adjustment to all tests simultaneously, there are also tests that apply adjustments in a stepwise manner, for example the Hochberg, Hommel and Holm tests. These tests have been shown to be powerful alternatives to the more traditional approaches, see Bate and Clark (2014) for more details.
Unadjusted tests
Fisher’s LSD (least significant difference)
Fisher’s LSD executes a series of t-tests among selected pairs of means, where the variability estimate used in all calculations is the more reproducible estimate taken from the ANOVA table (the Mean Square Error). This is one of the reasons why the LSD test is considered to be a more reliable test than simply performing lots of separate t-tests. However, it does not correct for multiple comparisons and it is thus associated with a higher false positive rate (type I error), making more likely to detect differences which are not real.
Number of factors of interest
Factorial designs (i.e. experiments with more than one factor of interest) are useful when the researcher has many different independent variables that may or may not influence the response and they wish to assess them all in one experiment. Running several experiments, where a single factor of interest is varied in each, might not make the most efficient use of the animals and does not allow interactions between the factors to be assessed. Modelling a condition often requires more than just one factor to explain changes in the outcome measure and factorial designs can be used to test the effect of multiple factors simultaneously.
Factorial designs, can be used to assess which levels of the independent variables of interest, such as sex, age and dose, should be selected to maximise the ‘window of opportunity’ to observe a treatment effect. For example, a study was conducted to assess an inflammatory response in dba vs. balb c strains of mice. This experiment allowed the scientist to select the strain that was most sensitive so that future experiments would have the largest window of opportunity when testing novel compounds and hence imply that the number of animals required could be reduced. When setting up a new animal model, while it may seem a waste of resources to run such pilot studies at the start of the experimental process, the long-term benefits can easily outweigh the initial costs.
Bate and Clark (2014) differentiate between two types of factorial design.
‘Large’ factorial designs are employed when the researcher wants to investigate the effect of many factors and how they interact with each other. The researcher may also want to identify those factors that can be ignored as having no significant effect on the response. These designs are particularly useful, for example, when setting up a new animal model. Large factorial designs necessarily involve many individual groups but because we are only interested in the overall effects, and do not need to make pairwise comparisons between the groups, the individual sample sizes can be small.
‘Small’ factorial designs, which are commonly applied in animal research, consist of usually no more than two or three factors. The purpose is to compare one group mean to another, using a suitable statistical test. Hence the experiment should be adequately powered (with a suitable sample sizes at each group) to allow pairwise comparisons between the combinations of factors.
References and further reading
ALTMAN, D. G. & BLAND, J. M. 2009. Parametric v non-parametric methods for data analysis. BMJ, 338, a3167.
BATE, S. T. & CLARK, R. A. 2014. The Design and Statistical Analysis of Animal Experiments, UK, Cambridge University Press.
BLAND, J. M. & ALTMAN, D. G. 1996. Transforming data. BMJ, 312, 770.
CONOVER, W. J. & IMAN, R. L. 1981. Rank Transformations as a Bridge between Parametric and Nonparametric Statistics. The American Statistician, 35, 124-129.
FESTING, M. F. & ALTMAN, D. G. 2002. Guidelines for the design and statistical analysis of experiments using laboratory animals. ILAR J, 43, 244-58.
MARINO, M. J. 2014. The use and misuse of statistical methodologies in pharmacology research. Biochem Pharmacol, 87, 78-92.
NAKAGAWA, S. & CUTHILL, I. C. 2007. Effect size, confidence interval and statistical significance: a practical guide for biologists. Biol Rev Camb Philos Soc, 82, 591-605.
http://www.3rs-reduction.co.uk/html/12__statisticanalysis.html
http://www.3rs-reduction.co.uk/html/11_regression.html
For more information of producing a normal probability plot: http://www.youtube.com/watch?v=1Ts2lYrXenE


